I was using the Ghose-Crippen ALogP predictor implemented in CDK as a QSAR for looking at peptide solubilities. Unfortunately, this partition coefficient predictor is fit on small organic molecules and does a pretty bad job at predicting peptide partition coefficients. Here's what it looks like on single amino acids compared with experimental data.
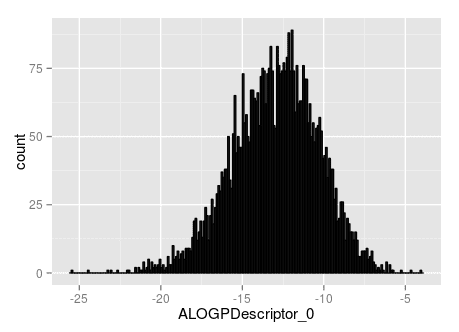
That's not too bad, all things considered. But, it gets much worse as the peptides get larger. Here's a histogram of the partition coefficients of a collection of 5,000 length 10 peptides.
As you can see, the partition coefficients are way too low. It turns out the partition coefficient predictions are correlated with the peptide length, but as someone who works with peptides, I know that their solubility is not a really a function of their length. Part of the problem for this is that the surface of these peptides is a linear function of weight because I'm not attempting to find the correct 3D geometry of the peptides.
How do I deal with this problem? Well, it has to do with my post on approximate integer partitions. I'm not sure if I'll have time to write it up; it'll definitely be in my next publication though! The basic idea is not to look at the absolute partition coefficient, but to compare them with similarly sized peptides. I can do that, because I'm using the predicted partition coefficient as a QSAR and am not actually trying to predict it.
That's not too bad, all things considered. But, it gets much worse as the peptides get larger. Here's a histogram of the partition coefficients of a collection of 5,000 length 10 peptides.
How do I deal with this problem? Well, it has to do with my post on approximate integer partitions. I'm not sure if I'll have time to write it up; it'll definitely be in my next publication though! The basic idea is not to look at the absolute partition coefficient, but to compare them with similarly sized peptides. I can do that, because I'm using the predicted partition coefficient as a QSAR and am not actually trying to predict it.